Ancient DNA reveals what really killed Napoleon's army
A fresh look at soldiers from the 1812 retreat out of Russia challenges the classic typhus explanation. Instead, ancient DNA pulled from teeth suggests enteric fever and relapsing fever were major disease players in the catastrophe that devastated Napoleon’s Grande Armée.
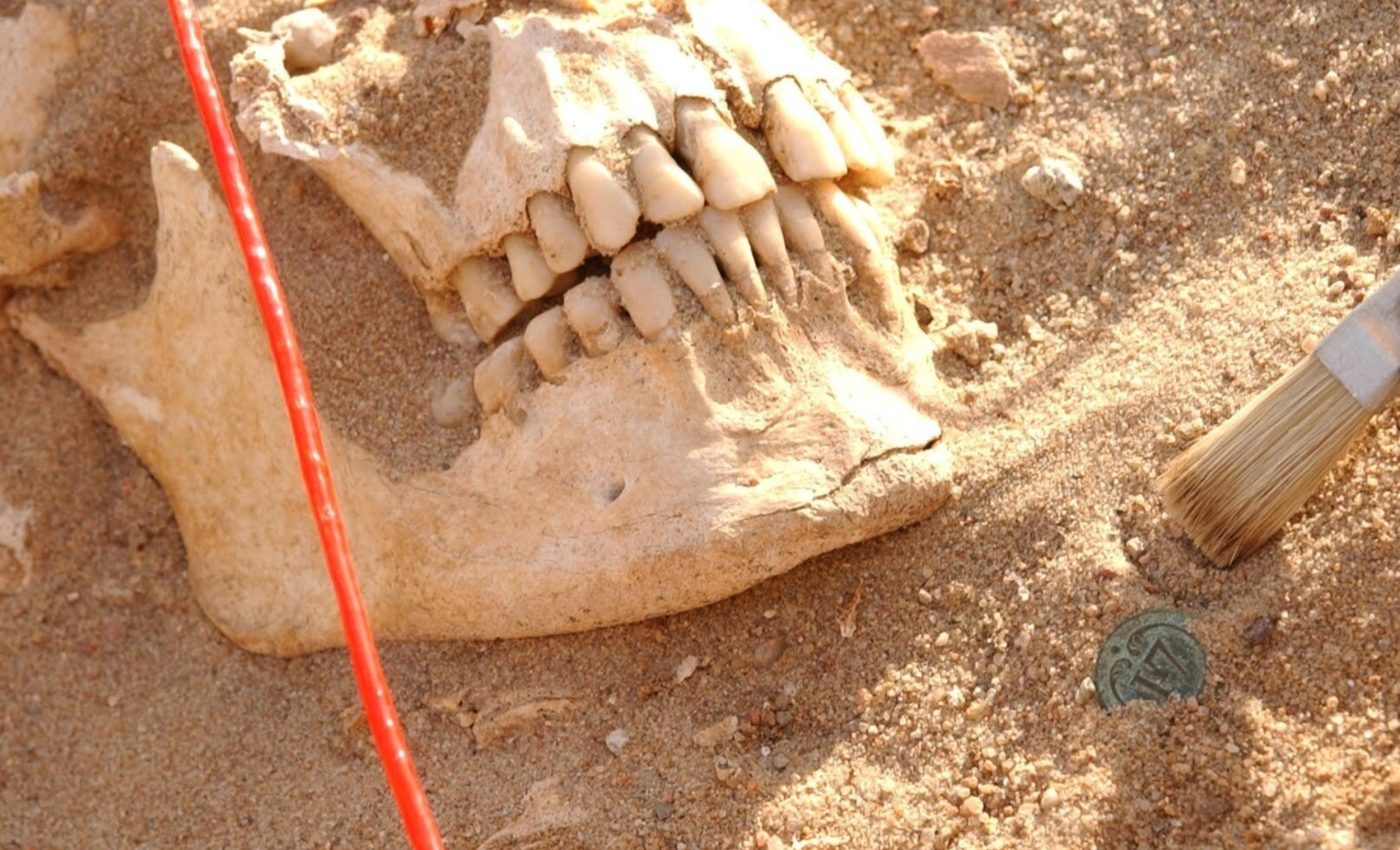
The study comes from a team of microbial paleogenomicists who reanalyzed remains from a mass grave on the army’s retreat route.
The authors include researchers from the Institut Pasteur in France. The team used modern sequencing to test whether typhus really drove the collapse – or whether other infections better fit the evidence.
Reexamining Napoleon’s frozen retreat
In the summer of 1812, Napoleon marched roughly half a million soldiers into the Russian Empire. By December, only a fraction remained alive.
Historians have long pointed to starvation, brutal cold, and epidemic disease for Napoleon’s collapse. Typhus became the shorthand culprit, supported by reports from doctors and officers of the time.
The discovery of body lice – typhus’s main vector – on remains from the campaign strengthened that case.
Even DNA from Rickettsia prowazekii, the bacterium behind epidemic typhus, had been reported from related material, keeping the typhus narrative alive.
DNA locked in enamel
The new study treated teeth as sealed archives. From 13 soldiers interred in a mass grave in Vilnius, Lithuania – on the path of the French retreat – the team extracted and sequenced DNA.
The challenge with centuries-old remains is contamination and decay. To solve this, the team applied filters to strip away environmental DNA and focus on authentic pathogen fragments.
The idea was simple: if typhus tore through the ranks, its genetic fingerprints should still be detectable.
DNA overturns the typhus tale
Instead of finding typhus, the researchers recovered traces of two other pathogens: Salmonella enterica, which causes enteric fever, and Borrelia recurrentis, the agent of louse-borne relapsing fever.
Both diseases match the conditions soldiers faced – crowding, poor sanitation, and malnutrition. Both can reach high mortality in stressed populations.
The team did not detect R. prowazekii or Bartonella quintana (the cause of trench fever), the latter of which has been reported in other studies of soldiers from the same site.
“It’s very exciting to use a technology we have today to detect and diagnose something that was buried for 200 years,” said lead author Nicolás Rascovan of the Institut Pasteur in France.
Old methods, missed signals
The discrepancy with past reports may come down to method. Earlier work often relied on PCR, a technique that amplifies a specific DNA segment. That approach can be powerful when the target segment is intact, but ancient DNA typically shatters into tiny pieces.
In contrast, broad, shotgun-style sequencing casts a wider net and can recover ultra-short fragments without needing a pristine, preselected target.
“Ancient DNA gets highly degraded into pieces that are too small for PCR to work. Our method is able to cast a wider net and capture a greater range of DNA sources based on these very short ancient sequences,” Rascovan explained.
A lineage that lingers
One surprise came from the relapsing fever bacterium. The B. recurrentis strain in these soldiers falls within the same lineage recently identified in ancient Britain, dating back about 2,000 years to the Iron Age.
That suggests a lineage that persisted across Europe for millennia before disappearing from the modern dataset. All present-day strains sequenced so far belong to a different branch.
The discovery shows how disease lineages rise, spread, and are replaced over long periods of time.
“This shows the power of ancient DNA technology to uncover the history of infectious diseases that we wouldn’t be able to reconstruct with modern samples,” Rascovan said.
Revisiting the Napoleon army disaster
None of this erases the lethal cold, hunger, and exhaustion that crushed Napoleon’s army. But it reframes the epidemic component.
Enteric fever, driven by S. enterica, thrives where sanitation collapses and clean water is scarce – conditions that surely prevailed during the chaotic retreat.
Louse-borne relapsing fever, carried by the same body lice that made typhus so feared, would have ridden on Napoleon’s soldiers’ clothing and spread rapidly through close contact, fueling disease across the ranks.
Together, these infections could have produced waves of fever, weakness, and death that compounded the winter’s toll.
The new data complicate a tidy narrative. The absence of R. prowazekii in this set of teeth suggests typhus may not have dominated these particular men’s final days, even if typhus circulated elsewhere or at other times during the campaign.
Sampling limits matter: thirteen individuals cannot stand in for an entire army scattered across thousands of kilometers and months of hardship. Still, the findings make it harder to treat typhus as the single, overarching explanation.
Tragic fate of Napoleon’s army
Beyond the 1812 story, the work shows how ancient DNA can revise received wisdom. Testing directly for pathogen DNA reduces reliance on inference from symptoms or vectors alone.
It also reveals hidden diversity – lineages that once were common but left no living descendants. Improving sequencing methods allows researchers to revisit more campaigns, sieges, and migrations from the historical record with greater precision.
The road home from Moscow wasn’t just a march through snow. Overcrowding, filth, and lice turned the camps into a gauntlet of infections.
In these 13 soldiers from Vilnius, the genetic trail leads to enteric fever and louse-borne relapsing fever, not the classic epidemic typhus.
That shift doesn’t lessen the tragedy – it sharpens it. Different pathogens, same brutal outcome. And a reminder that history’s most famous disasters can still yield new answers when we let the dead speak through their DNA.
Image credit: Michel Signoli, Aix-Marseille Université
The full study is published in the journal Current Biology.
—–
Like what you read? Subscribe to our newsletter for engaging articles, exclusive content, and the latest updates.
Check us out on EarthSnap, a free app brought to you by Eric Ralls and Earth.com.
—–













