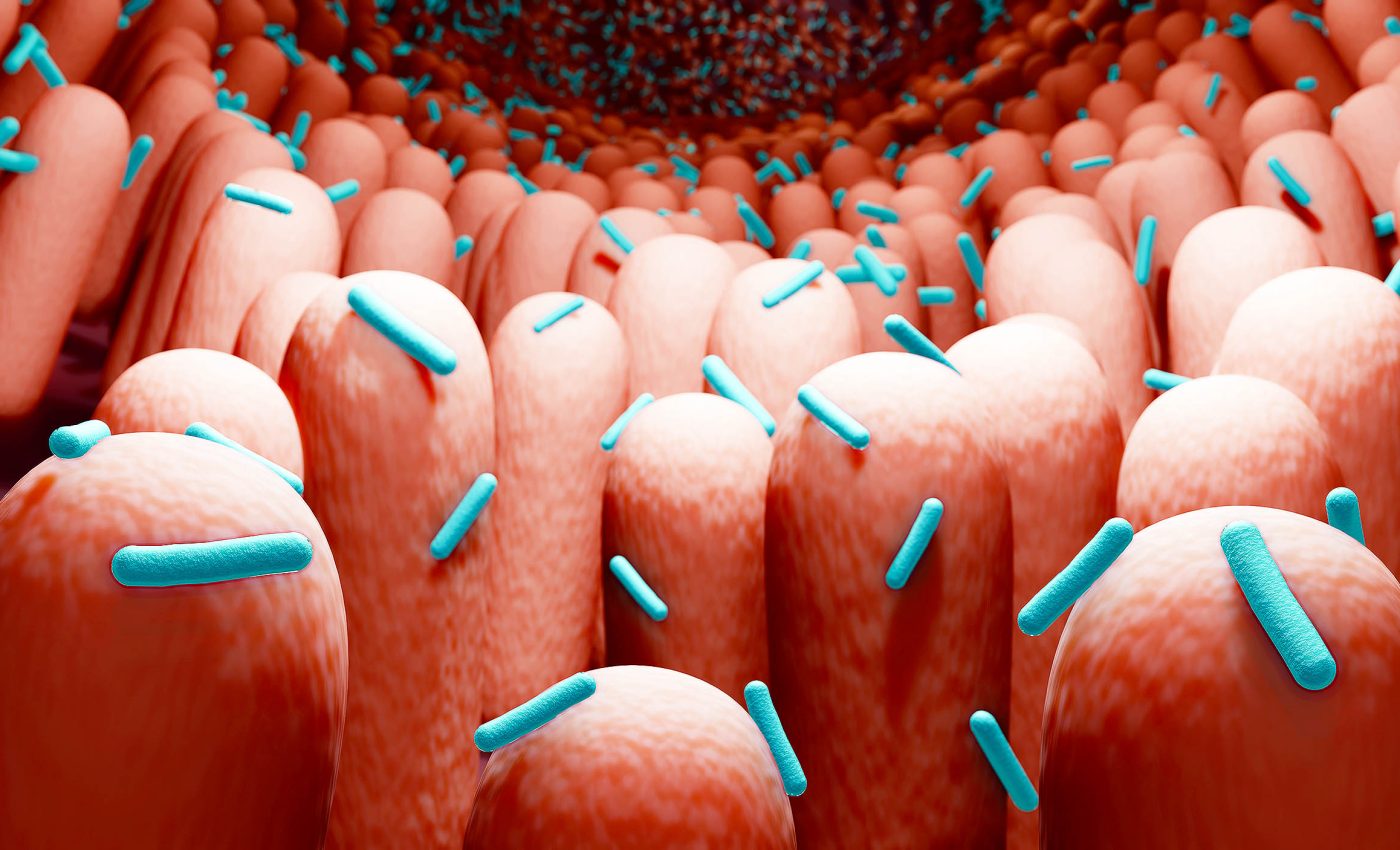
New life forms found in the human body, discovery described by scientists as 'insane'
Humans carry around a busy community of microbes that help run daily life. This microbiome breaks down food, trains the immune system, and competes with germs.
Scientists just spotted something unexpected inside those communities: tiny circular RNAs that don’t match any known virus or bacterium.
These RNAs appear in human-associated samples, especially from the mouth, and they look and act in ways that set them apart from other known living entities in the human body
The work points to a layer of biology that has been present in our data all along, but somehow overlooked.
It wasn’t found with a new microscope or a specialized culture method. It was found by reading patterns in large RNA datasets and asking careful questions about structure and sequence.
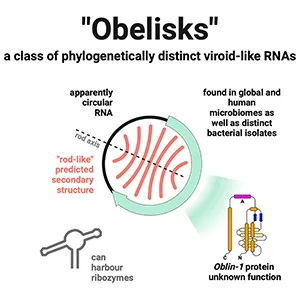
The result is a new category of RNA the team calls “obelisks.”
Understanding RNA obelisks
Obelisks are small loops of RNA, each roughly a thousand genetic “letters” long. Folding predictions show a long, rod-like shape with repeated base-pairing along the length. That shape echoes certain plant pathogens called viroids, but the match isn’t perfect.
Unlike classic plant viroids, some obelisks carry a stretch of RNA that appears to encode a protein. The group nicknamed those putative proteins “Oblins.”
This mix – circular RNA that forms a rod and sometimes encodes a protein – doesn’t fit the usual categories. That difference is the key fact – viroids don’t encode proteins.
Many typical RNA viruses do encode proteins but package their genomes in different ways. Obelisks cut a new path.
How to find RNA obelisks
The team sifted through metatranscriptomic data, which is a pooled set of RNA from entire microbial communities. Their software screened for two hallmarks of circular RNAs.
One hallmark is the telltale assembly overlap where the end meets the beginning. The other is a predicted rod-shaped hairpin produced by RNA folding models.
By requiring both signals and then tightening filters, the pipeline pulled out a robust set of candidates from tens of thousands of initial hits.
This approach shows how algorithms can work as discovery tools when the rules of the search are defined by biochemistry. Build the right test for a circle and a rod-like fold, and unfamiliar RNAs rise out of the noise.
Signals for obelisks appeared in many human-associated datasets, but the mouth stood out. Oral RNA samples carried obelisks in a large fraction of cases, with fewer matches in the gut.
One person’s oral samples showed the same obelisk signatures persisting for many months. That pattern looks consistent with stable colonization rather than random fragments passing through.
The simplest reading is that the oral microbiome offers hosts or conditions that suit these RNAs. Saliva, biofilms on teeth, and the mix of common oral bacteria could set the stage for maintenance over time.
Clues about replication
Several obelisks carry a short sequence motif known as a hammerhead ribozyme. A ribozyme is RNA that performs a chemical task without a protein helper.
Hammerhead ribozymes can cut and rejoin RNA strands, steps that fit a replication cycle for circular molecules. Having built-in “scissors” would let a small RNA manage part of its own copying machinery.
Not every obelisk carries this motif. The presence across several examples, though, points to a self-processing strategy in at least part of the group.
That shifts the picture from “weird pieces of RNA” toward evolved mechanisms that have adapted to survive and multiply.
Bacteria as RNA obelisk hosts
The data point to common mouth bacteria as likely hosts. One case stands out: Streptococcus sanguinis, a common oral species.
Researchers saw a specific obelisk at high levels in the bacterial RNA but could not find matching DNA in the bacterial genome.
That mismatch supports the idea that the obelisk is a separate RNA replicating alongside the bacterium rather than a gene encoded in its chromosome.
In lab culture, the bacterium grew without obvious problems in the absence of the obelisk. That outcome suggests the obelisk is not essential for survival in standard conditions, yet its steady presence in natural samples implies a real biological relationship.
Why does any of this matter?
Obelisks blur lines that textbooks try very hard to keep sharp. They are viroid-like in size and folding but sometimes encode a protein. RNA obelisks also show up in human-associated microbial communities rather than plants.
Finally, they persist across time and carry motifs that hint at self-processing replication. All of this expands what counts as a “genetic entity.”
These features broaden how we think about microbial ecology. If obelisks can replicate in or alongside common bacteria, they may add subtle layers of interaction – competition for enzymes, changes in transcription, or tweaks to stress responses.
Human health implications
There is no evidence that obelisks cause disease in humans. The responsible reading is that they mark a newly recognized layer of the microbiome’s complexity.
They may act as subtle influencers of bacterial behavior, or they may ride along with little effect. Both ideas are testable.
Clear next steps sit on the bench: culture host microbes with and without their obelisks, watch replication in controlled experiments, map how any encoded Oblin proteins function, and measure changes in growth, metabolism, or interactions with neighboring microbes.
Those tests will connect sequence features to real-world outcomes.
Lessons from the RNA obelisk discovery
Sequencing now captures vast amounts of genetic material from oceans, soils, and the human body. Hidden in those reads are patterns that fail to match existing categories.
When searches are built on first principles – like circular assembly signatures and stable RNA folds – new classes of biology can come into view. Obelisks are a case where careful algorithm rules surfaced entities that standard methods overlooked.
This lesson travels well. Similar pipelines could scan for other unusual structures, different catalytic motifs, or hybrid features that mix traits we usually treat as separate.
The more we look with targeted tests, the more likely we are to find biology that doesn’t follow our old boundaries.
The full study was published in the journal Cell.
—–
Like what you read? Subscribe to our newsletter for engaging articles, exclusive content, and the latest updates.
Check us out on EarthSnap, a free app brought to you by Eric Ralls and Earth.com.
—–













